Data wrangling#
Goals of this lecture#
A huge \(\%\) of science involves data wrangling.
This could be an entire course on its own, but today we’ll focus on:
What is data wrangling?
What to do about missing values?
How to combine datasets?
Tidy data!
What is it?
How do we make our data tidy?
Importing relevant libraries#
import seaborn as sns ### importing seaborn
import pandas as pd
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
What is data wrangling?#
Data wrangling refers to manipulating, reshaping, or transforming a dataset as needed for your goals (e.g., visualization and/or analysis).
A huge \(\%\) of working with data involves “wrangling”.
Includes:
“Cleaning” data (missing values, recasting variables, etc.).
Merging/combining different datasets.
Reshaping data as needed.
Dealing with missing values#
In practice, real-world data is often messy.
This includes missing values, which take on the value/label
NaN.NaN= “Not a Number”.
Dealing with
NaNvalues is one of the main challenges in CSS!
Loading a dataset with missing values#
The titanic dataset contains information about different Titanic passengers and whether they Survived (1 vs. 0).
Commonly used as a tutorial for machine learning, regression, and data wrangling.
df_titanic = pd.read_csv("data/wrangling/titanic.csv")
df_titanic.head(3)
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 2 | 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
Why is missing data a problem?#
If you’re unaware of missing data:
You might be overestimating the size of your dataset.
You might be biasing the results of a visualization or analysis (if missing data are non-randomly distributed).
You might be complicating an analysis.
By default, many analysis packages will “drop” missing data––so you need to be aware of whether this is happening.
How to deal with missing data#
Identify whether and where your data has missing values.
Analyze how these missing values are distributed.
Decide how to handle them.
Not an easy problem––especially step 3!
Step 1: Identifying missing values#
The first step is identifying whether and where your data has missing values.
There are several approaches to this:
Using
.isnaUsing
.infoUsing
.isnull
isna()#
The
isna()function tells us whether a given cell of aDataFramehas a missing value or not (Truevs.False).If we call
isna().any(), it tells us which columns have missing values.
df_titanic.isna().head(1)
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | False | False | False | False | False | False | False | False | False | False | True | False |
df_titanic.isna().any()
PassengerId False
Survived False
Pclass False
Name False
Sex False
Age True
SibSp False
Parch False
Ticket False
Fare False
Cabin True
Embarked True
dtype: bool
Inspecting columns with nan#
Now we can inspect specific columns that have nan values.
df_titanic[df_titanic['Age'].isna()].head(5)
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 0 | 3 | Moran, Mr. James | male | NaN | 0 | 0 | 330877 | 8.4583 | NaN | Q |
| 17 | 18 | 1 | 2 | Williams, Mr. Charles Eugene | male | NaN | 0 | 0 | 244373 | 13.0000 | NaN | S |
| 19 | 20 | 1 | 3 | Masselmani, Mrs. Fatima | female | NaN | 0 | 0 | 2649 | 7.2250 | NaN | C |
| 26 | 27 | 0 | 3 | Emir, Mr. Farred Chehab | male | NaN | 0 | 0 | 2631 | 7.2250 | NaN | C |
| 28 | 29 | 1 | 3 | O'Dwyer, Miss. Ellen "Nellie" | female | NaN | 0 | 0 | 330959 | 7.8792 | NaN | Q |
How many nan?#
If we call sum on the nan values, we can calculate exactly how many nan values are in each column.
df_titanic.isna().sum()
PassengerId 0
Survived 0
Pclass 0
Name 0
Sex 0
Age 177
SibSp 0
Parch 0
Ticket 0
Fare 0
Cabin 687
Embarked 2
dtype: int64
info#
The info() function gives us various information about the DataFrame, including the number of not-null (i.e., non-missing) values in each column.
df_titanic.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 891 entries, 0 to 890
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PassengerId 891 non-null int64
1 Survived 891 non-null int64
2 Pclass 891 non-null int64
3 Name 891 non-null object
4 Sex 891 non-null object
5 Age 714 non-null float64
6 SibSp 891 non-null int64
7 Parch 891 non-null int64
8 Ticket 891 non-null object
9 Fare 891 non-null float64
10 Cabin 204 non-null object
11 Embarked 889 non-null object
dtypes: float64(2), int64(5), object(5)
memory usage: 83.7+ KB
Check-in#
How many rows of the DataFrame have missing values for Cabin?
### Your code here
Solution#
### How many? (Quite a few!)
df_titanic[df_titanic['Cabin'].isna()].shape
(687, 12)
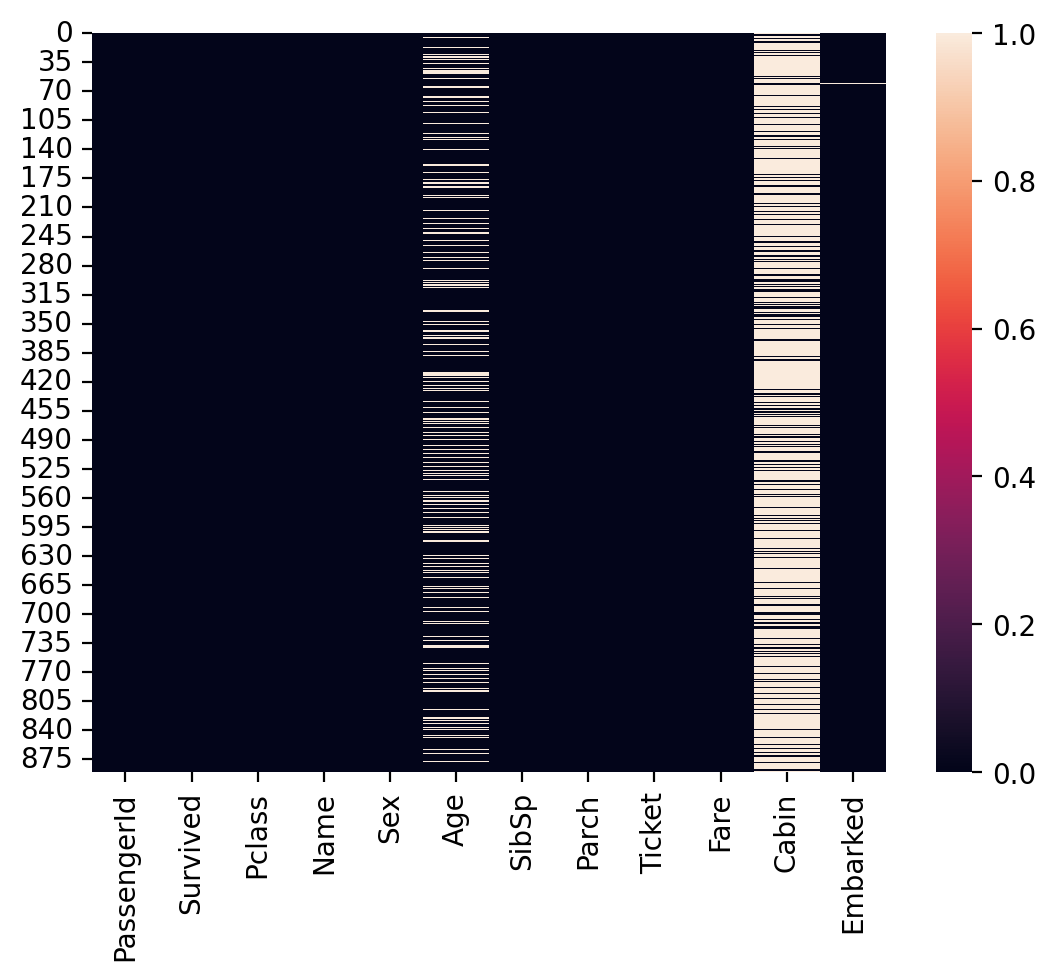
Visualizing missing values#
Finally, we can visualize the rate of missing values across columns using
seaborn.heatmap.The dark cells are those with not-null values.
The light cells have
nanvalues.
sns.heatmap(df_titanic.isna())
<Axes: >
Step 2: Analyze how the data are distributed#
Having identified missing data, the next step is determining how those missing data are distributed.
Is variable \(Y\) different depending on whether \(X\) is nan?#
One approach is to ask whether some variable of interest (e.g., Survived) is different depending on whether some other variable is nan.
### Mean survival for people without data about Cabin info
df_titanic[df_titanic['Cabin'].isna()]['Survived'].mean()
0.29985443959243085
### Mean survival for people *with* data about Cabin info
df_titanic[~df_titanic['Cabin'].isna()]['Survived'].mean()
0.6666666666666666
Check-in#
What is the mean Survived rate for values with a nan value for Age vs. those with not-null values? How does this compare to the overall Survived rate?
### Your code here
Solution#
### Mean survival for people without data about Age
df_titanic[df_titanic['Age'].isna()]['Survived'].mean()
0.2937853107344633
### Mean survival for people *with* data about Age
df_titanic[~df_titanic['Age'].isna()]['Survived'].mean()
0.4061624649859944
### Mean survival for people overall
df_titanic['Survived'].mean()
0.3838383838383838
Additional methods#
If you’re interested in diving deeper, you can look into the missingno library in Python (which must be separately installed).
Step 3: Determine what to do!#
Having identified missing data, you need to determine how to handle it.
There are several approaches you can take.
Removing all rows with any missing data.
Removing rows with missing data only when that variable is relevant to the analysis or visualization.
Imputing (i.e., guessing) what values missing data should have.
Removing all rows with any missing data#
We can filter our
DataFrameusingdropna, which will automatically “drop” any rows containing null values.Caution: if you have lots of missing data, this can substantially impact the size of your dataset.
df_filtered = df_titanic.dropna()
df_filtered.shape
(183, 12)
Removing all rows with missing data in specific columns#
Here, we specify that we only want to
dropnafor rows that havenanin theAgecolumn specifically.We still have missing
nanforCabin, but perhaps that’s fine in our case.
df_filtered = df_titanic.dropna(subset = "Age")
df_filtered.shape
(714, 12)
Imputing missing data#
One of the most complex (and controversial) approaches is to impute the values of missing data.
There are (again) multiple ways to do this:
Decide on a constant value and assign it to all
nanvalues.E.g., assign the
meanAgeto all people withnanin that column.
Try to guess the value based on specific characteristics of the data.
E.g., based on other characteristics of this person, what is their likely
Age?
Imputing a constant value#
We can use fillna to assign all values with nan for Age some other value.
## Assign the mean Age to all people with nan for Age
df_titanic['Age_imputed1'] = df_titanic['Age'].fillna(df_titanic['Age'].mean())
## Now let's look at those rows
df_titanic[df_titanic['Age'].isna()].head(5)
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Age_imputed1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 0 | 3 | Moran, Mr. James | male | NaN | 0 | 0 | 330877 | 8.4583 | NaN | Q | 29.699118 |
| 17 | 18 | 1 | 2 | Williams, Mr. Charles Eugene | male | NaN | 0 | 0 | 244373 | 13.0000 | NaN | S | 29.699118 |
| 19 | 20 | 1 | 3 | Masselmani, Mrs. Fatima | female | NaN | 0 | 0 | 2649 | 7.2250 | NaN | C | 29.699118 |
| 26 | 27 | 0 | 3 | Emir, Mr. Farred Chehab | male | NaN | 0 | 0 | 2631 | 7.2250 | NaN | C | 29.699118 |
| 28 | 29 | 1 | 3 | O'Dwyer, Miss. Ellen "Nellie" | female | NaN | 0 | 0 | 330959 | 7.8792 | NaN | Q | 29.699118 |
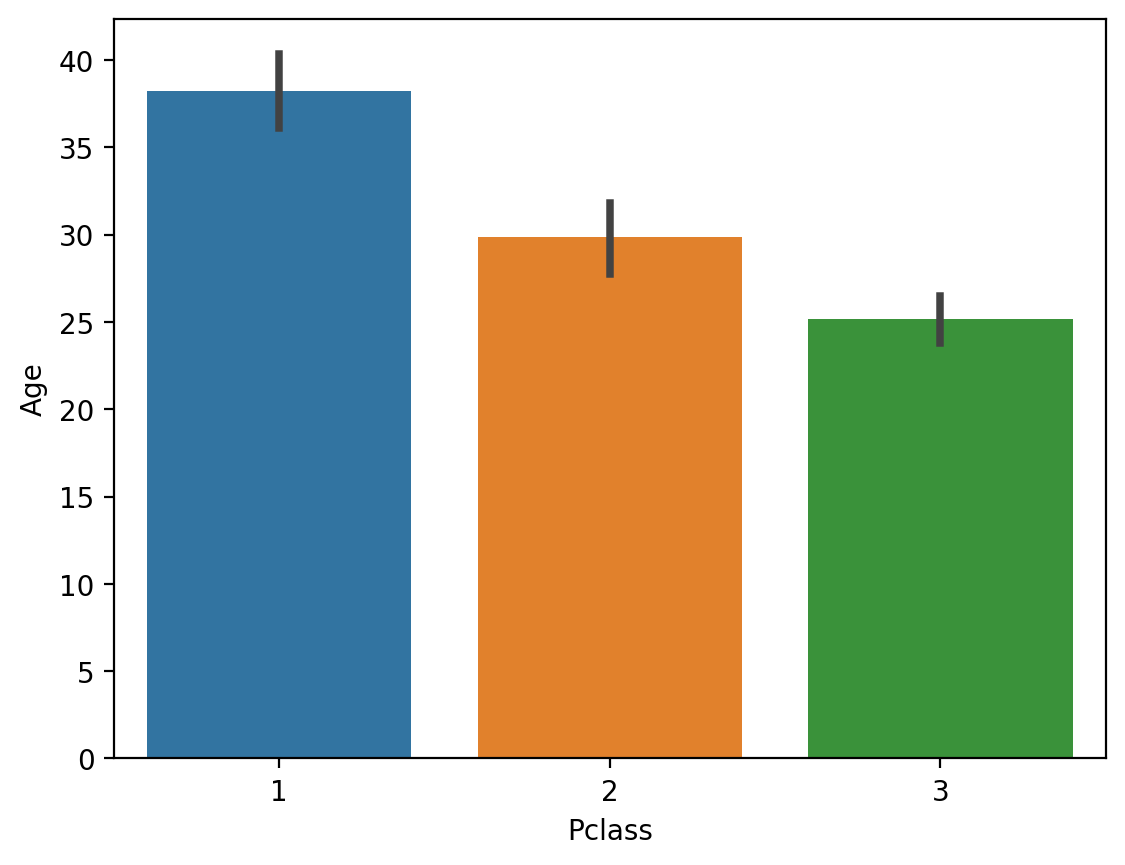
Guessing based on other characteristics#
You can try to guess what their
Agewould be, based on other features.The more sophisticated version of this is to use statistical modeling or using
SimpleImputerfrom thesklearnlibrary.For now, simply note that
Agecorrelates with other features (likePclass).
## Passenger Class is correlated with Age
sns.barplot(data = df_titanic, x = 'Pclass', y = 'Age')
<Axes: xlabel='Pclass', ylabel='Age'>
Check-in#
What would happen if you used fillna with the median Age instead of the mean? Why would this matter?
### Your code here
Solution#
The median Age is slightly lower.
## Assign the median Age to all people with nan for Age
df_titanic['Age_imputed2'] = df_titanic['Age'].fillna(df_titanic['Age'].median())
## Now let's look at those rows
df_titanic[df_titanic['Age'].isna()].head(5)
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Age_imputed1 | Age_imputed2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 0 | 3 | Moran, Mr. James | male | NaN | 0 | 0 | 330877 | 8.4583 | NaN | Q | 29.699118 | 28.0 |
| 17 | 18 | 1 | 2 | Williams, Mr. Charles Eugene | male | NaN | 0 | 0 | 244373 | 13.0000 | NaN | S | 29.699118 | 28.0 |
| 19 | 20 | 1 | 3 | Masselmani, Mrs. Fatima | female | NaN | 0 | 0 | 2649 | 7.2250 | NaN | C | 29.699118 | 28.0 |
| 26 | 27 | 0 | 3 | Emir, Mr. Farred Chehab | male | NaN | 0 | 0 | 2631 | 7.2250 | NaN | C | 29.699118 | 28.0 |
| 28 | 29 | 1 | 3 | O'Dwyer, Miss. Ellen "Nellie" | female | NaN | 0 | 0 | 330959 | 7.8792 | NaN | Q | 29.699118 | 28.0 |
Merging datasets#
Merging refers to combining different datasets to leverage the power of additional information.
In Week 1, we discussed this in the context of data linkage.
Can link datasets as a function of:
Shared time window.
Shared identity.
Why merge?#
Each dataset contains limited information.
E.g.,
GDPbyYear.
But merging datasets allows us to see how more variables relate and interact.
Much of social science research involves locating datasets and figuring out how to combine them.
How to merge?#
In Python, pandas.merge allows us to merge two DataFrames on a common column(s).
pd.merge(df1, df2, on = "shared_column")
merge in practice#
For demonstration, we’ll merge two Linguistics datasets:
One dataset contains information about the Age of Acquisition of different English words (Kuperman et al., 2014).
The other dataset contains information about the Frequency and Concreteness of English words (Brysbaert et al., 2014).
Loading datasets#
df_aoa = pd.read_csv("data/wrangling/AoA.csv")
df_aoa.head(1)
| Word | AoA | |
|---|---|---|
| 0 | a | 2.89 |
df_conc = pd.read_csv("data/wrangling/concreteness.csv")
df_conc.head(1)
| Word | Concreteness | Frequency | Dom_Pos | |
|---|---|---|---|---|
| 0 | sled | 5.0 | 149 | Adjective |
Different kinds of merging#
As we see, the datasets are not the same size. This leaves us with a decision to make when merging.
innerjoin: Do we preserve only the words in both datasets?leftjoin: Do we preserve all the words in one dataset (the “left” one), regardless of whether they occur in the other?rightjoin: Do we preserve all the words in one dataset (the “right” one), regardless of whether they occur in the other?outerjoin: Do we preserve all words in both, leaving empty (nan) values where a word only appears in one dataset?
df_aoa.shape
(31124, 2)
df_conc.shape
(28612, 4)
inner join#
For our purposes, it makes the most sense to use an
innerjoin.This leaves us with fewer words than occur in either dataset.
df_merged = pd.merge(df_aoa, df_conc, on = "Word", how = "inner")
df_merged.head(2)
| Word | AoA | Concreteness | Frequency | Dom_Pos | |
|---|---|---|---|---|---|
| 0 | aardvark | 9.89 | 4.68 | 21 | Noun |
| 1 | abacus | 8.69 | 4.52 | 12 | Noun |
df_merged.shape
(23569, 5)
Check-in#
What happens if you use a different kind of join, e.g., outer or left? What do you notice about the shape of the resulting DataFrame? Do some rows have nan values?
### Your code here
Solution#
df_outer_join = pd.merge(df_aoa, df_conc, on = "Word", how = "outer")
df_outer_join.shape
(36167, 5)
df_outer_join.head(4)
| Word | AoA | Concreteness | Frequency | Dom_Pos | |
|---|---|---|---|---|---|
| 0 | a | 2.89 | NaN | NaN | NaN |
| 1 | aardvark | 9.89 | 4.68 | 21.0 | Noun |
| 2 | abacus | 8.69 | 4.52 | 12.0 | Noun |
| 3 | abalone | 12.23 | NaN | NaN | NaN |
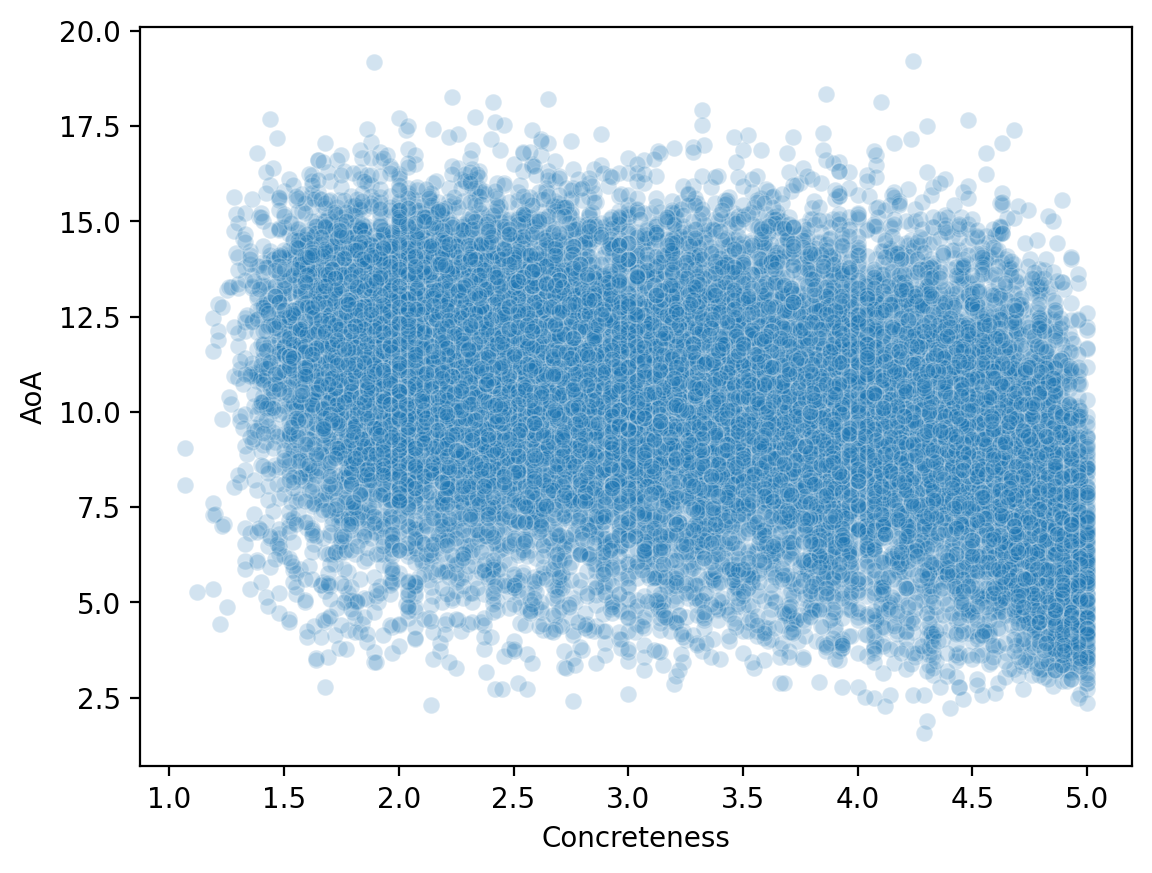
Why merge is so useful#
Now that we’ve merged our datasets, we can look at how variables across them relate to each other.
sns.scatterplot(data = df_merged, x = 'Concreteness',
y = 'AoA', alpha = .2 )
<Axes: xlabel='Concreteness', ylabel='AoA'>
Reshaping data#
Reshaping data involves transforming it from one format (e.g., “wide”) to another format (e.g., “long”), to make it more amenable to visualization and analysis.
Often, we need to make our data tidy.
What is tidy data?#
Tidy data is a particular way of formatting data, in which:
Each variable forms a column (e.g.,
GDP).Each observation forms a row (e.g., a
country).Each type of observational unit forms a table (tabular data!).
Originally developed by Hadley Wickham, creator of the tidyverse in R.
Tidy vs. “untidy” data#
Now let’s see some examples of tidy vs. untidy data.
Keep in mind:
These datasets all contain the same information, just in different formats.
“Untidy” data can be useful for other things, e.g., presenting in a paper.
The key goal of tidy data is that each row represents an observation.
Tidy data#
Check-in: Why is this data considered tidy?
df_tidy = pd.read_csv("data/wrangling/tidy.csv")
df_tidy
| ppt | condition | rt | |
|---|---|---|---|
| 0 | john | Congruent | 200 |
| 1 | john | Incongruent | 250 |
| 2 | mary | Congruent | 178 |
| 3 | mary | Incongruent | 195 |
Untidy data 1#
Check-in: Why is this data not considered tidy?
df_messy1 = pd.read_csv("data/wrangling/messy1.csv")
df_messy1
| john | mary | condition | |
|---|---|---|---|
| 0 | 200 | 178 | Congruent |
| 1 | 250 | 195 | Incongruent |
Untidy data 2#
Check-in: Why is this data not considered tidy?
df_messy2 = pd.read_csv("data/wrangling/messy2.csv")
df_messy2
| congruent | incongruent | ppt | |
|---|---|---|---|
| 0 | 200 | 250 | john |
| 1 | 178 | 195 | mary |
Making data tidy#
Fortunately, pandas makes it possible to turn an “untidy” DataFrame into a tidy one.
The key function here is pandas.melt.
pd.melt(df, ### Dataframe
id_vars = [...], ### what are the identifying columns?
var_name = ..., ### name for variable grouping over columns
value_name = ..., ### name for the value this variable takes on
If this seems abstract, don’t worry––it’ll become clearer with examples!
Using pd.melt#
Let’s start with our first messy
DataFrame.Has columns for each
ppt, which contain info aboutrt.
df_messy1
| john | mary | condition | |
|---|---|---|---|
| 0 | 200 | 178 | Congruent |
| 1 | 250 | 195 | Incongruent |
pd.melt(df_messy1, id_vars = 'condition', ### condition is our ID variable
var_name = 'ppt', ### new row for each ppt observation
value_name = 'rt') ### label for the info we have about each ppt
| condition | ppt | rt | |
|---|---|---|---|
| 0 | Congruent | john | 200 |
| 1 | Incongruent | john | 250 |
| 2 | Congruent | mary | 178 |
| 3 | Incongruent | mary | 195 |
Check-in#
Try to use pd.melt to turn df_messy2 into a tidy DataFrame.
Hint: Think about the existing structure of the DataFrame––how is data grouped––and what the id_vars would be.
df_messy2
| congruent | incongruent | ppt | |
|---|---|---|---|
| 0 | 200 | 250 | john |
| 1 | 178 | 195 | mary |
### Your code here
Solution#
pd.melt(df_messy2, id_vars = 'ppt', ### here, ppt is our ID variable
var_name = 'condition', ### new row for each ppt observation
value_name = 'rt') ### label for the info we have about each ppt
| ppt | condition | rt | |
|---|---|---|---|
| 0 | john | congruent | 200 |
| 1 | mary | congruent | 178 |
| 2 | john | incongruent | 250 |
| 3 | mary | incongruent | 195 |
Hands-on: a real dataset#
Now, we’ll turn to a real dataset, which Timothy Lee, creator of Full Stack Economics, compiled and shared with me.
df_work = pd.read_csv("data/viz/missing_work.csv")
df_work.head(5)
| Year | Child care problems | Maternity or paternity leave | Other family or personal obligations | Illness or injury | Vacation | Month | |
|---|---|---|---|---|---|---|---|
| 0 | 2012 | 18 | 313 | 246 | 899 | 1701 | 10 |
| 1 | 2012 | 35 | 278 | 230 | 880 | 1299 | 11 |
| 2 | 2012 | 13 | 245 | 246 | 944 | 1005 | 12 |
| 3 | 2013 | 14 | 257 | 250 | 1202 | 1552 | 1 |
| 4 | 2013 | 27 | 258 | 276 | 1079 | 1305 | 2 |
Check-in#
Is this dataset tidy? How could we make it tidy, if not––i.e., if we wanted each row to be a single observation corresponding to one of the reasons for missing work?
### Your code here
Solution#
df_melted = pd.melt(df_work, id_vars = ['Year', 'Month'],
var_name = "Reason",
value_name = "Days Missed")
df_melted.head(2)
| Year | Month | Reason | Days Missed | |
|---|---|---|---|---|
| 0 | 2012 | 10 | Child care problems | 18 |
| 1 | 2012 | 11 | Child care problems | 35 |
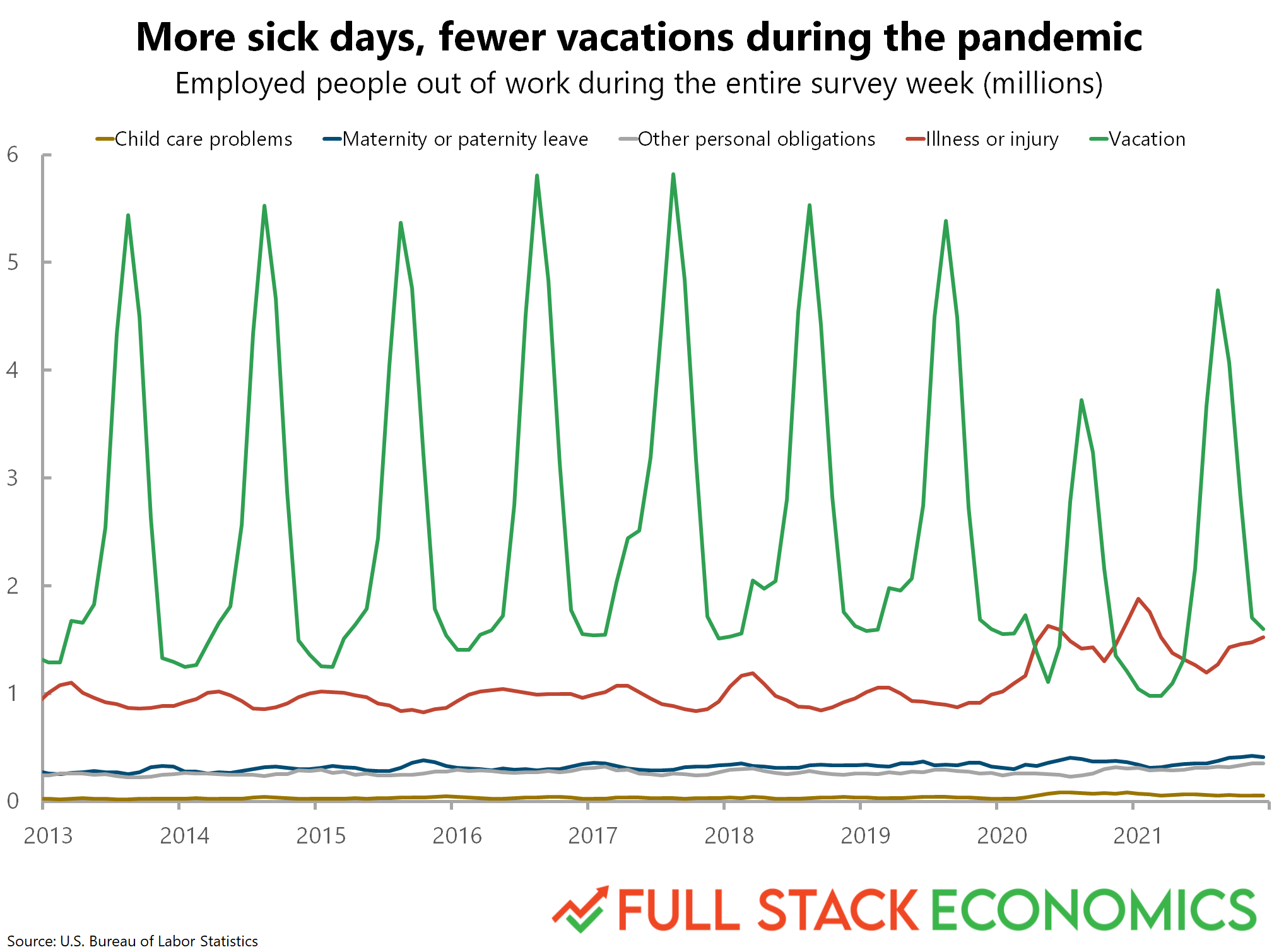
Why tidy data is useful#
Finally, let’s use this dataset to recreate a graph from FullStackEconomics.
Original graph#
Check-in#
As a first-pass approach, what tools from seaborn could you use to recreate this plot?
### Your code here
Solution#
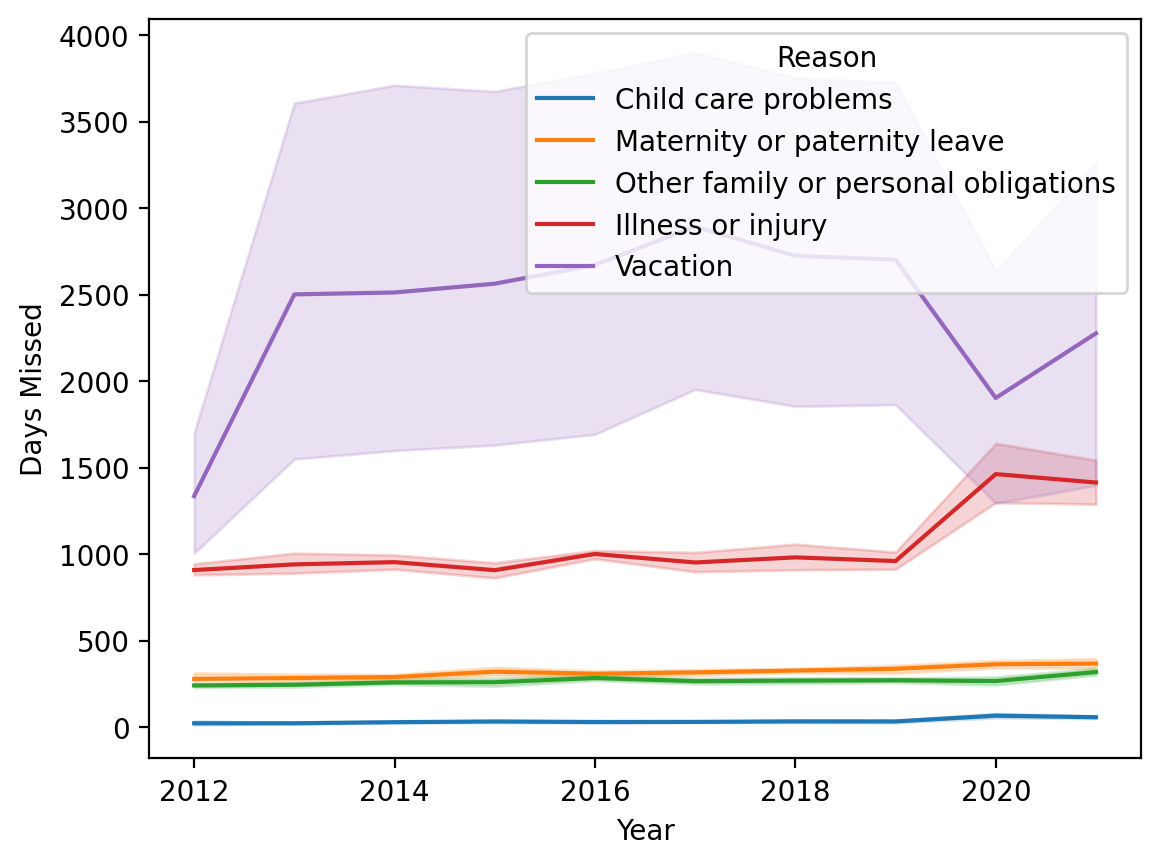
This is okay, but not really what we want. This is grouping it by Year. But we want to group by both Year and Month.
sns.lineplot(data = df_melted, x = 'Year', y = 'Days Missed', hue = "Reason")
<Axes: xlabel='Year', ylabel='Days Missed'>
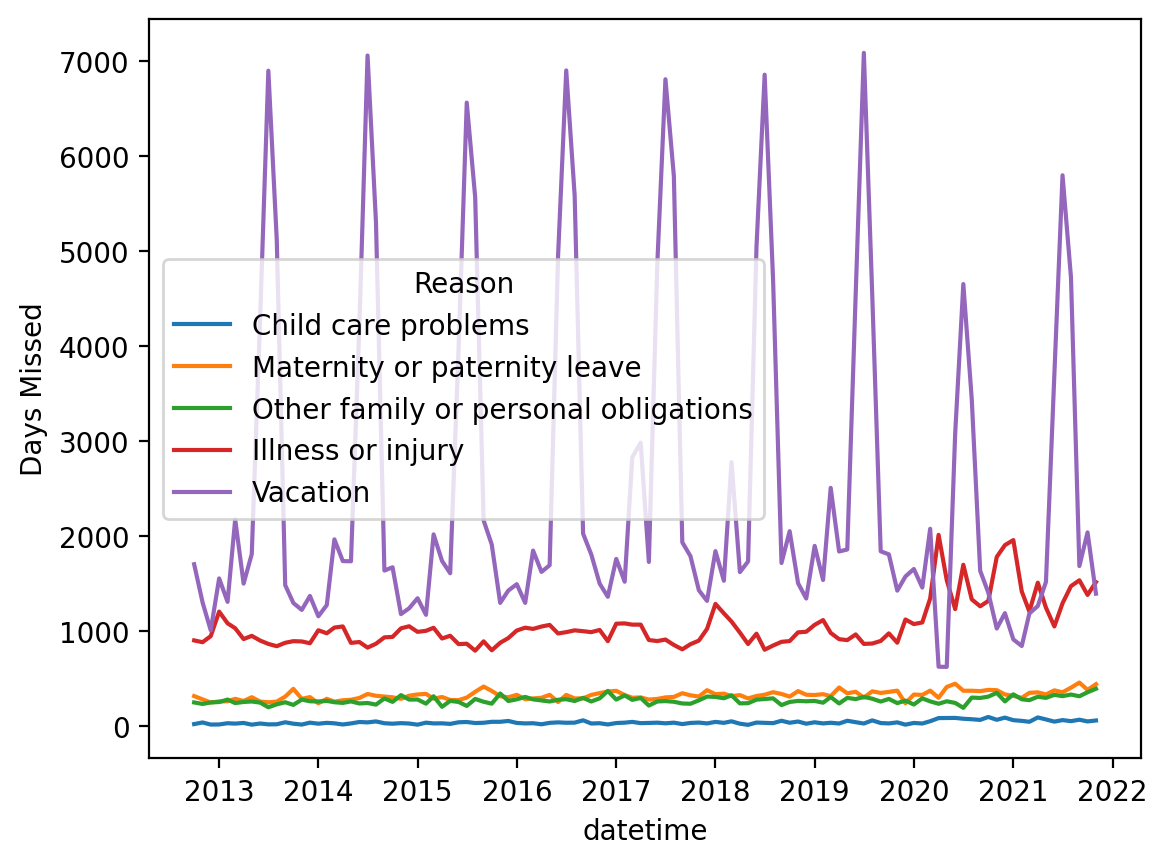
Using datetime#
Let’s make a new column called
date, which combines theMonthandYear.Then we can use
pd.to_datetimeto turn that into a custompandasrepresentation.
## First, let's concatenate each month and year into a single string
df_melted['date'] = df_melted.apply(lambda row: str(row['Month']) + '-' + str(row['Year']), axis = 1)
df_melted.head(2)
| Year | Month | Reason | Days Missed | date | |
|---|---|---|---|---|---|
| 0 | 2012 | 10 | Child care problems | 18 | 10-2012 |
| 1 | 2012 | 11 | Child care problems | 35 | 11-2012 |
## Now, let's create a new "datetime" column using the `pd.to_datetime` function
df_melted['datetime'] = pd.to_datetime(df_melted['date'])
df_melted.head(2)
/var/folders/pn/5zbmv0cj31v6hmyh53njhmdw0000gn/T/ipykernel_1691/714386078.py:2: UserWarning: Could not infer format, so each element will be parsed individually, falling back to `dateutil`. To ensure parsing is consistent and as-expected, please specify a format.
df_melted['datetime'] = pd.to_datetime(df_melted['date'])
| Year | Month | Reason | Days Missed | date | datetime | |
|---|---|---|---|---|---|---|
| 0 | 2012 | 10 | Child care problems | 18 | 10-2012 | 2012-10-01 |
| 1 | 2012 | 11 | Child care problems | 35 | 11-2012 | 2012-11-01 |
Plotting again#
Much better!
sns.lineplot(data = df_melted, x = "datetime", y = "Days Missed", hue = "Reason")
<Axes: xlabel='datetime', ylabel='Days Missed'>
Conclusion#
This was an introduction to data wrangling.
As noted, data wrangling is a hugely important topic––and could constitute an entire class.
But today, we’ve focused on:
Identifying and addressing missing data.
Merging datasets.
Making data tidy.
